Prostate cancer (PCa) is a complex disease with diverse cancer cell types, each contributing to its heterogeneity. The cancer genome’s genomic instability and epigenetic mechanisms drive clonal competition and cellular differentiation, leading to the emergence of various PCa cell subtypes. Among them, prostate cancer stem cells (PCSCs) play a crucial role, sharing similarities with normal stem cells. Since their discovery in 2005, researchers have been tirelessly exploring the unique metabolic characteristics of PCSCs, delving into the biological mysteries of this intriguing aspect of prostate cancer.
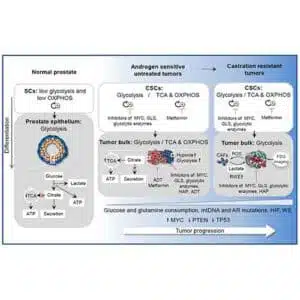
Prostate cancer (PCa) is heterogeneous, harboring phenotypically diverse cancer cell types. Genomic instability leads to clonal competition and evolution of the cancer genome, and epigenetic mechanisms result in subclonal cellular differentiation, resulting in PCa cell heterogeneity. Tumor cell differentiation begins with a population of prostate cancer stem cells (PCSCs), which share many phenotypic and functional characteristics with normal stem cells. Since the first publications on PCSCs in 2005, researchers have worked hard to understand their biological aspects, especially their distinct metabolic characteristics.
Prostate cancer is a highly heterogeneous disease, and understanding the biological aspects of prostate cancer stem cells (PCSCs) has been the focus of researchers since their discovery in 2005. PCSCs share many characteristics with normal stem cells and are responsible for clonal tumor evolution. This article discusses the link between the tumor initiating cells and CSCs that maintain tumor growth, and how metastases are driven by evolved populations of CSCs. Additionally, the article explores the potential use of CSC analysis as a prognostic or predictive biomarker for tailored therapy selection.
Prostate cancer development is an evolutionary process that reflects an evolving of CSCs. Luminal or basal cells in normal prostate can act as a cell of tumor origin after oncogenic transformation. The link between the tumor initiating cells, or tumor cell-of-origin and CSCs that maintain tumor growth is not yet understood. Cancer cell-of-origin refers to the initial normal cell or the cell type that became tumorigenically
transformed whereas CSCs refer to the cell population that drives clonal tumor evolution. Tumor metastases are driven by the evolved populations of CSCs at their worst. Some tumor cells with malignant potential enter the blood stream (CTCs) and can be disseminated to the distant organs. Single prostate tumor cells can be found in bone marrow and are called DTCs. Single tumor cells disseminated to lymph nodes are called isolated tumor cells (ITCs, not shown). A small subset of DTCs or ITCs could give rise to metastasis – initiating cells (MICs). Metastatic spread and formation is a long-time process that might take a few years.
Once developed metastases can form secondary metastasis to distant organs. Prostate cancer progression is associated with development of substantial intra-tumor heterogeneity and genomic instability that can be induced by MYC activation, loss of PTEN and mutations in DNA repair genes including BRCA2, ATM, and CHEK2. A few PCSC phenotypes have been validated so far in animal models for their tumor initiating properties, for example, PSA−/lo, ALDHhigh, NANOG+, Trop2+, CD44+CD133+, ALDHhighCD44+α2β1+.
PCSCs with self-renewal characteristics and genomic instability could be a driving force behind tumor progression and metastatic spread. CSC analysis in patients’ tumors has been shown in a number of recent studies to be a prognostic or predictive biomarker. These markers could be employed in the future for patient categorization and more tailored therapy selection when combined with other extrinsic and intrinsic parameters that may alter the features and complexity of CSC populations, such as heterogeneity indices and hypoxia.
© 2025 Immucura. All Rights Reserved
Loading: Please wait…
Loading form: Please wait…
Loading: Please wait…
Loading form… Please wait…
Loading form: please wait…